What is Bayesian Linear Regression? (Part 1)
Matthias Werner
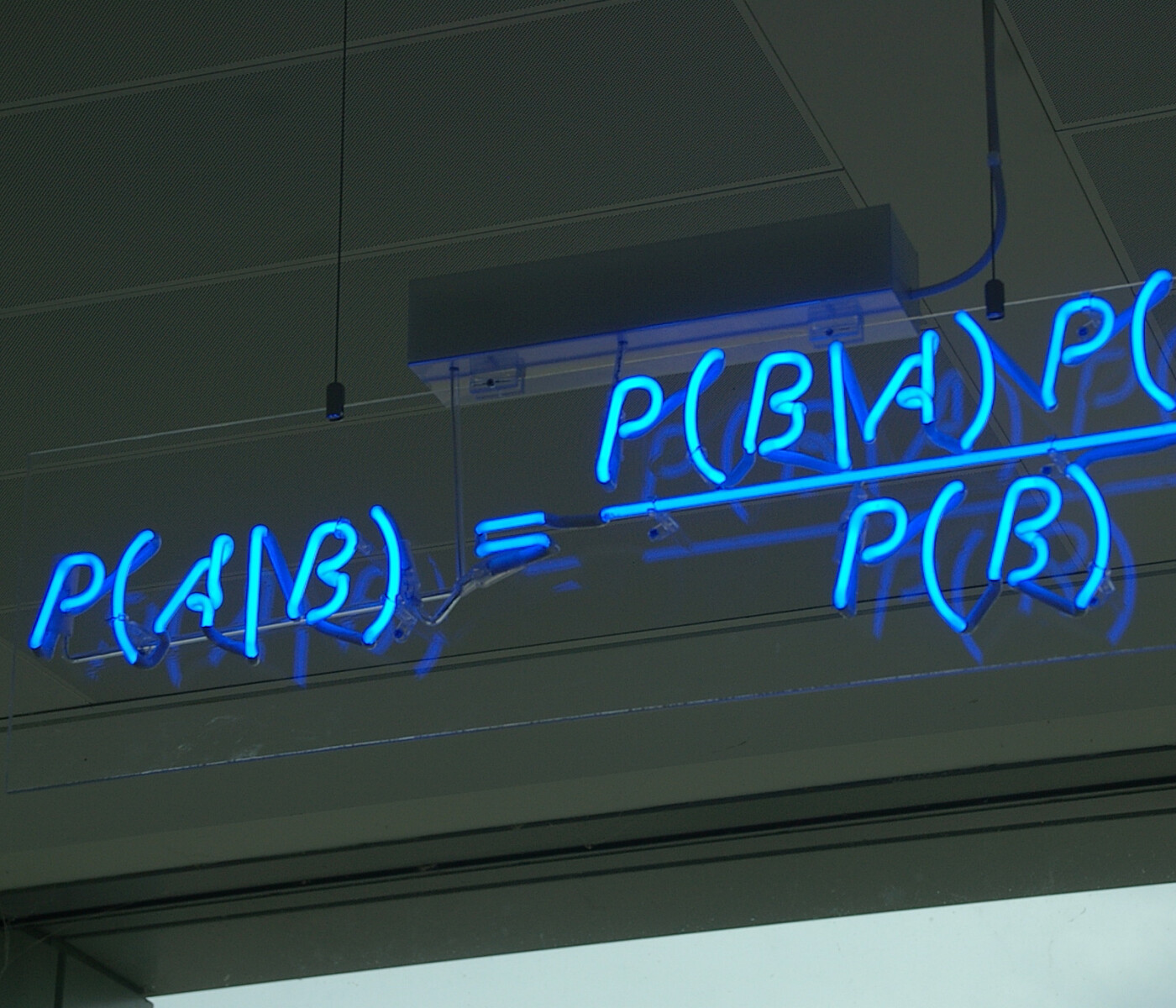
Bayesian regression methods are very powerful, as they not only provide us with point estimates of regression parameters, but rather deliver an entire distribution over these parameters. This can be understood as not only learning one model, but an entire family of models and giving them different weights according to their likelihood of being correct. As this weight distribution depends on the observed data, Bayesian methods can give us an uncertainty quantification of our predictions representing what the model was able to learn from the data. The uncertainty measure could be e.g. the standard deviation of the predictions of all the models, something that point estimators will not provide by default. Knowing what the model doesn't know helps to make AI more explainable. To clarify the basic idea of Bayesian regression, we will stick to discussing Bayesian Linear Regression (BLR).
BLR is the Bayesian approach to linear regression analysis. We will start with an example to motivate the method. To make things clearer, we will then introduce a couple of non-Bayesian methods that the reader might already be familiar with and discuss how they relate to Bayesian regression.
In the following I assume that you have elementary knowledge of linear algebra and stochastics.
Let's get started!
A simple example
The task we want to solve is the following: Assume we are given a set $$D$$ of $$N_D$$ independently generated, noisy data points $$D := \{ (x_i, y_i)\}_{i=1}^{N_D}$$, where $$x_i \in \mathbb{R}^m$$ and $$y_i \in \mathbb{R}^n$$ (in our example $$m=n=1$$). We know that there is some relationship between the $$x_i$$ and the $$y_i$$, but we do not know what this relationship is. We will make a simplifying assumption, namely that the relationship is linear, but we also have some measurement noise on our hands. Formally, we can write this as
where for convenience we define $$\vec{x}_i := (1, x_i)^T$$ and $$\vec{w} := (w_0, w_1)^T$$. $$w_0$$ and $$w_1$$ are the model parameters that we want to estimate. All $$\epsilon_i$$ are independently drawn from a normal distribution with standard deviation $$\sigma_\epsilon$$. We do not want to model the noise $$\epsilon_i$$, but rather use it as an excuse that our model will not represent the data perfectly.
Assume we already know the posterior distribution $$p(\vec{w}|D)$$ which encodes what we think the parameters $$\vec{w}$$ could be after observing the data $$D$$ (we will learn how to obtain it in a minute). For a given input $$\vec{x}$$ we get an entire distribution over the predictions $$y = \vec{w}^T \vec{x}$$ weighted by $$p(\vec{w}|D)$$. This distribution is called the predictive distribution.
We can visualize the predictive distribution by plotting the mean and the standard deviation of the prediction for a given $$x$$. The neat thing about BLR is that we can compute these moments analytically. For the mean prediction we get
where we use the linearity of both expectation value and scalar product. For the variance we need to compute
where $$\vec{\mu}_w$$ and $$\Sigma_w$$ are the mean vector and the covariance matrix of $$p(\vec{w}|D)$$.
Furthermore, we can also sample $$p(\vec{w}|D)$$ and plot the resulting models. As $$\Sigma_w$$ is positive-definite (we'll assume this to be true without proving it here, but it has something to do with the fact that it is the sum of a positive semi-definite matrix and an identity matrix), we can decompose it such that
If we draw a vector $$\vec{z}$$ from a multi-variate standard Gaussian
we can convert it into a sample of the posterior using
Before observing data we simply draw from the prior distribution $$p(\vec{w}) = \mathcal{N}(\vec{0}, I)$$ (where $$I$$ is the identity matrix), which we choose to be a multi-variate standard Gaussian. Then we gradually introduce new data points, compute the posterior and plot the distribution and the sampled models.

We can see that without observing data points, we predict on average a zero, but with a large variance. As we observe more and more data points, the mean prediction converges and the standard deviation decreases. Intuitively this makes perfect sense, as we become more and more certain of our predictions as the number of observed data increases.
Now let's find out about the math behind BLR.
Bayes' Theorem
As the name BLR already suggests, we will make use of Bayes' theorem. In particular, we will use the formulation of Bayes' theorem for probability densities. The probability densities $$p_X$$, $$p_{X|Y}$$ and $$p_{XY}$$ are defined such that
for some $$a \leq b$$ and $$c \leq d$$. Bayes' theorem for probability densities states that
In some sense Bayes' theorem allows us to "flip" the conditional probabilities, all we need are the unconditional distributions. Within the Bayesian framework, the unconditional distributions are called the prior distributions, or priors in short, while the conditional distributions are called the posteriors. In practice, the "Bayesian" workflow looks as follows:
We want to compute $$p_{Y|X}(y|x)$$, but only have access to $$p_{X|Y}(x|y)$$.
Therefore we need to come up with a prior $$p_Y(y)$$ which should represent our prior "belief" about $$y$$.
Now we can compute the product $$p_{X|Y}(x|y) p_Y(y)$$ which for a given $$x$$ is some (hopefully integrable) function of $$y$$.
To turn an integrable function of $$y$$ into a probability density, it needs to be normalized via some constant $$N$$ (which might depend on $$x$$) such that $$\int p_{Y|X}(y|x) d_y = \frac{1}{N} \int p_{X|Y}(x|y) p_Y(y) dy = 1$$, where the integration is done over all possible $$y$$.
The last point implies
which is the denominator of Bayes' Theorem. In short, if we wanted to compute $$p_{Y|X}(y|x)$$, but only had access to $$p_{X|Y}(x|y)$$, often times we would just define a prior $$p_Y(y)$$ and obtain $$p_{Y|X}(y|x)$$ by normalizing the numerator of Bayes' Theorem. Computing the normalization can be quite costly at times, so this laziness might fall on ones feet, unless steps are taken to mitigate the intractability of the normalization integral in the formula above (e.g. variational inference).
Next, let us look at non-Bayesian linear regression in more detail and discuss how it relates to the Bayesian counter-part.
Linear Regression
Maximum Likelihood Estimator
If you have ever solved a small (or sometimes even a big) regression problem you most likely used an Maximum Likelihood Estimator (MLE). The likelihood of the model parameters is defined as
Conveniently, as the noise is additive, we obtain a conditional distribution
Our goal is now to find the set of model parameters $$\vec{w}_{MLE}^{\ast}$$ that maximizes the likelihood as defined above. For convenience we often use the log-likelihood $$LL$$. As the logarithm is strictly increasing, taking the logarithm of the likelihood does not change the location of the maximum $$\vec{w}_{MLE}^{\ast}$$. This gives
The last step follows since $$p(y_i | \vec{x}_i, \vec{w})$$ is a Gaussian probability density. The terms with the logarithm in the summation do not depend on the model parameters $$\vec{w}$$, therefore they can be ignored in the optimization. As scaling the optimization objective by a positive number does not change the location of the optimum either, we can furthermore omit the factor $$\frac{1}{2\sigma^2_\epsilon}$$ in the first term. This gives the optimization problem
This is a neat result. The MLE for a linear model under the assumption of additive Gaussian noise turns out to minimize the sum of squared errors.
Maximum A-Posteriori
The MLE is already quite nice, sometimes, however, it might be advantageous to incorporate prior assumptions about the model parameters, e.g. when we only have a small amount of noisy data. This can be done using the Maximum A-Posteriori estimator estimator (MAP). For MLE we tried to find the set of parameters $$\vec{w}$$ such that the probability of the observed data $$p(D|\vec{w})$$ is maximized. An alternative approach would be to maximize the probability $$p(\vec{w}|D)$$ of the parameters having observed the data $$D$$. As $$p(\vec{w}|D)$$ is the posterior distribution over the parameters, this is where MAP gets its name from, i.e. we want to find the model parameters $$\vec{w}^{\ast}_{MAP}$$ that maximizes the posterior distribution given the data $D$ we observed. But what does $$p(\vec{w}|D)$$ look like?
Let's consider Bayes' theorem one more time:
The optimization problem we need to solve now can be written as
As the $$p(yi_|\vec{x}_i)$$ do not depend on the model parameters $$\vec{w}$$, they end up as simple factors that do not influence the location of $$\vec{w}^{\ast}_{MAP}$$, therefore we can drop them. Furthermore, we can again use the monotony of the logarithm to write
As we can see, the summation over the data points is the same as in the MLE, we have simply introduced one additional term $$\log p(\vec{w})$$ which allows us to formulate prior beliefs about the model parameters.
For a quick example let's assume our parameters to be distributed normally and independently around the origin with a variance $$\sigma^2_w$$, i.e.
Throwing together the last two equations with the conditional distribution derived above, we get
where we again drop parameter-independent terms. Since we can scale the optimization objective with any positive scalar value we want and are basically free to choose $$\sigma_w^2$$ for our prior beliefs, we can rewrite the optimization objective as
We see that MAP solves almost the same optimization problem as MLE with an extra additional term scaled by some parameter $$\lambda$$ for which we need to find a reasonable value. $$\lambda$$ basically quantifies how strongly we belief that the model parameters are close to zero. Readers with some knowledge in Machine Learning will recognize that MAP is the same as MLE with L2-regularization.
Bayesian Linear Regression
We have just used Bayes' theorem to justify estimating the model parameters with regularization, but we are still using point estimates of the model parameters $$\vec{w}$$. Bayes' theorem could theoretically give us access not just to the maximum of the posterior distribution as in MAP, but allow us to use the distribution $$p(\vec{w} | D)$$ itself. This will allow us to estimate the standard deviation of our prediction. We will use conjugate priors to compute $$p(\vec{w} | D)$$. Conjugate priors are prior distributions $$p(\vec{w})$$ which are in the same family of distributions as the posterior $$p(\vec{w} | D)$$. As in the examples above, we will assume the likelihood $$p(y_i | \vec{x}_i, \vec{w})$$ and the prior $$p(\vec{w})$$ to be Gaussians. A Gaussian times a Gaussian is still a Gaussian. As it can be seen in Bayes' Theorem, the formula for normalization and Bayes' Theorem, the prior of the training data $$p(D)$$ is essentially a normalization constant and will not influence the general shape of $$p(\vec{w} | D)$$. So we already know $$p(\vec{w} | D)$$ will be a Gaussian. For a Gaussian we only need to figure out the mean vector $$\vec{\mu}_w$$ and the covariance matrix $$\Sigma_w$$ of $$p(\vec{w} | D)$$ and then can infer the normalization from there.
That's it for now. In the second part I will explain the details of the math behind Bayesian Linear Regression.
